On Apr 10th, 2018, Dr. ZHU Bing’s lab at the Institute of Biophysics (IBP), Chinese Academy of Sciences (CAS) published a paper entitled “Roles of the CSE1L-mediated nuclear import pathway in epigenetic silencing” in PNAS. In this study, they described a regulator, CSE1L, that is essential for the silencing of many endogenous methylated genes, and proposed that the cargo specificity of the protein nuclear import system may impact the selectivity of gene silencing.
DNA methylation mediated gene silencing contributes to various biological processes, including transcription regulation, X chromosome inactivation, genomic imprinting, and transposon silencing. Known mechanisms involved in this process were direct inhibition of certain transcription factors, histone deacetylation by histone deacetylases (HDACs) recruited by methyl-CpG binding proteins (MBDs), recruitment of site-specific repressors, histone H3K9 methylation, and chromatin condensation mediated by other chromatin factors.
To identify novel regulators involved in DNA methylation-mediated gene silencing, the authors devised a cell-based reporter system and used an unbiased RNAi screening to identify regulators involved in this process. They identified CSE1L, a key player previously known in the nuclear import pathway, as an essential factor for maintaining the repression of many methylated genes. Depletion of CSE1L reactivates the DNA methylation silenced reporter gene and many endogenous methylated genes. These depressed genes largely overlapped with methylated genes that were also reactivated by treatment with histone deacetylase inhibitors (HDACi). Mechanically, CSE1L depletion did not cause DNA demethylation and gene silencing defects observed upon CSE1L depletion were linked to its nuclear import function for certain protein cargos. Certain nuclear-localized silencing factors, including NOVA1, HDAC1, HDAC2, and HDAC8, became delocalized into cytosol upon CSE1L depletion. And, depletion of other factors involved in the same import nuclear pathway, such as KPNAs and KPNB1, also displayed similar depression profiles at the genome-wide level. This study showed that the cargo specificity of the protein nuclear import system may impact the selectivity of methylated gene silencing, which advance the understanding of DNA methylation-related gene silencing.
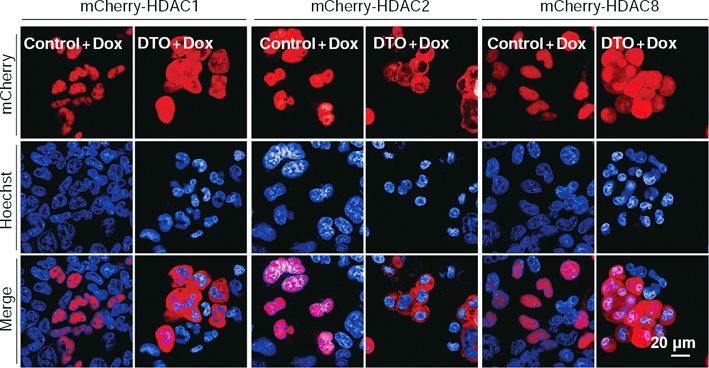
CSE1L depletion selectively inhibits the nuclear localization of certain transcription repressors, such as mCherry-HDAC1, mCherry-HDAC2, and mCherry-HDAC8 (DONG Qiang et al., PNAS, 2018)
Dr. ZHU Bing and Dr. ZHANG Zhuqiang are co-corresponding authors. DONG Qiang from ZHU Bing’s group is the first author. Dr. WANG Hailing ’s group from Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences; and Dr. LEI Xiaoguang’s group from Peking University participated in this study. This work was supported by the National Natural Science Foundation of China, the Chinese Ministry of Science and Technology, and CAS.
Article link: http://www.pnas.org/content/early/2018/04/09/1800505115
Contact:
Dr. ZHU Bing
Email: zhubing@ibp.ac.cn
Institute of Biophysics, CAS

